Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Analysis of the Expression of SELL, ADCY8 and TLR4 in Low Grade Gliomas and Correlation with Prognosis Based on Tumor Subtype Clustering
*Corresponding author: Shaoping_fan, People’s Hospital of Longhua, Shenzhen, China.
Received: December 06, 2023; Published: December 13, 2023
DOI: 10.34297/AJBSR.2023.20.002768
Abstract
Purpose: To identify a specific group with poor prognosis in LGG(low-grade glioma), analyze the expression of SELL,ADCY8 and TLR4 in LGG, associated pathways and their association with patient outcomes.
Material and Methods: Bioinformatics analysis ConsensusClusterPlus cluster analysis was used to identify specific groups with poor prognosis in LGG. By the GSEA algorithm, significantly up-regulated pathway was found. Furthermore, in the TCGA database dataset and CGGA database data, it was found that there were prognostic related genes SELL, ADCY8 and TLR4, high expression was significantly associated with poor prognosis in LGG.
Results: In TCGA and CGGA database data, there was a specific group in the LGG database, the prognosis of this group was worse than that of other group (three groups are identified by clustering algorithm), the up-regulated genes in this group are mainly involved in immune cell interaction, immune cell migration, antigen presentation, cytokine pathway and other related pathways. The upregulated genes SELL, ADCY8 and TLR4 in EC1 were lower than that of patients with low expression of TLR4 (SELL p.Value=0.0435, ADCY8 p.value=0.0435, TLR4 p.value=0.0367).
Conclusions: There is a specific group in LGG that has a significantly poor prognosis and the significantly up-regulated tumor immune-related pathway compared with other groups. The high expression of SELL, ADCY8 and TLR4 in LGG were associated with poor prognosis and immune infiltration.
Keywords: Prognostic subgroups, Low-grade glioma, Prognostic genes, Tumor immunity
Abbreviations: LGG: Low Grade Glioma; SELL: Sell Selectins; ADCY8 and TLR4: Toll Receptor-Like 4; GSEA: Gene Set Enrichment Analysis; TIMER: Tumor Immune Estimation Resource
Introduction
In neurosurgery, Low-Grade Gliomas (LGG) is the most common primary tumor in the central nervous system, according to the latest CBTRUS report, LGG accounts for 15% of primary CNS tumors [1], although low-grade gliomas grow slowly, however, some LGG are difficult to resect completely and tend to degenerate into glioblastoma, leading to a poor consequence [2]. At present, a large number of studies combined with medical big data have demon strated that tumor immune abnormalities are closely related to LGG progression [3-6]. Therefore, basic research is needed to further clarify the relationship between tumor immunity and the relationship between LGG progression and the clinical need for an accurate and simple LGG adjuvant prognostic indicators.
Sell selectins are members of the selectin family (the family also includes selectins-P. Selectin and selectin E), often expressed on the surface of immune cells involved in the infiltration of immune cells, are expressed in many tumors, Resto and komiluk, et al. found that selectin plays an important role in leukocyte migration and infiltration, and in the tumor immune microenvironment TLR4 (Toll receptor-like 4), in immune related pathway NF-kappa B [7]. It has been found that ADCY8 is highly expression in LGG, and ADCY8 is regulation gene in the CAMP pathway and lead to gene FN1 abnormal activation in cAMP pathway and increases proliferation of glioma cells [8,9], these results suggest that SELL, ADCY8 and TLR4 play an important role in tumor immunity and tumorigenesis of LGG. However, the expression and mechanism of the above three genes in LGG have not been reported, and their value in the diagnosis of LGG is not clear.
Material and Methods
LGG gene expression matrix in TCGA and CCGA (expression matrix has been normalized, batch effect removed, and transformed into TPM expression pattern), a total of 959 patients with LGG. Bioinformatics Consensus ClusterPlus clustering method [10]. A very poor prognostic cohort was found in 515 LGG patients in the TCGA database. Discovery of GSEA Method in bioinformatics. This extremely poor prognostic group expressed up-regulated genes and related functional groups. Kaplan-Meier survival analysis was used to analyze the prognostic genes, and P < 0.05 was used as the index of difference. TIMER online tool to analyze gene association with tumor infiltration.
Results
We first selected a gene expression matrix from 515 low-grade gliomas, a prognostic range group was found in TCGA LGG by bioinformatics Consensus ClusterPlus clustering (Figure 1) by comparing it to the IDH mutation, most of this group is IDH wild type, but some samples of this group are not DH wild type, on the contrary, there is no obvious overlap between this group and a pathological type of LGG (Figure 2).
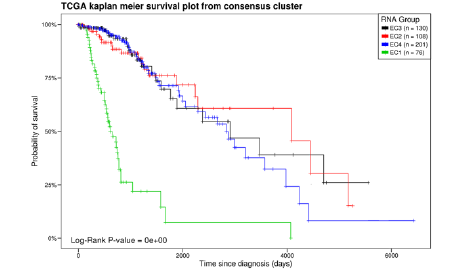
Figure 1: Kaplan-meier Survival Curve. EC1-EC4 is the biological information analysis cluster to get four clusters of which EC1 cluster is significantly worse than the other three groups.

Figure 2: Heat map of LGC gene expression: Heat map of BC1 - ECI gene expression, in which BC1 and IDH were significantly repopulated.
The EC1 Genes in the Poor Prognosis Group are Mainly Enriching in the Tumor Immune Pathway
By using the GSEA bioinformatics analysis method, it was found that EC1 compared with the other three groups EC2-EC4, the upregulated genes were mainly concentrated in immune cell interaction, immune cell migration, immune cell antigen presentation, cytokine pathway and other related pathways, these pathways play an important role in tumor immune infiltration (Figure 3).
The expression of SELL, ADCY8 and TLR4 are Closely Related to the Prognosis of LGG
In order to assess the relationship between the up-regulated gene of this prognostic group EC1 (the composition of the up-regulated pathway for GSEA analysis) in LGG, we concluded that Kaplan- meier curves combined with analysis (high expression and low expression by genes above and below their mean expression values) found that SELL, ADCY8 and TLR4 are prognostic genes in EC1 and LGG prognostic-related genes (Figure 4).
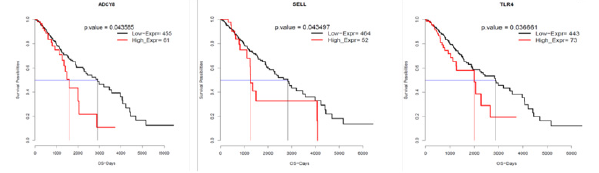
Figure 4: Kapln-meier survival analysis. Red and brown lines represent high and low expression, respectively.
SELL, ADCY8 and TLR4 were Associated with Multiple Immune Functions of LGG
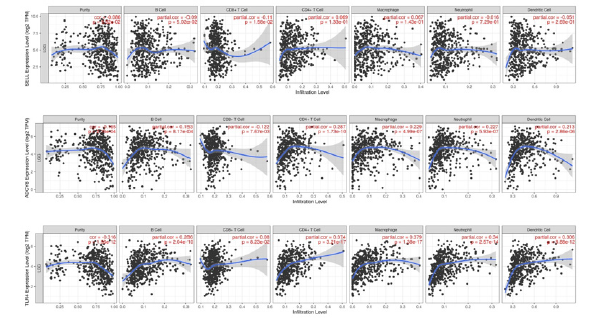
Through online tumor immune cell infiltration analysis tool TIMER (Tunor Innume Estination Resource, Https://cistrome.shinyapps. io/tiner/), It was found that SELL, ADCY8 and TLR4 were correlated with the infiltration of various immune functions (Figure 5).
Discussion
In recent years, a large number of studies have demonstrated that LGG tumor-immune response plays an important role in tumorigenesis and progression [12-15]. For example, we can see various degrees of infiltration of workers’ lymphocytes in glioma specimens excised from wood, in addition, the immune infiltration status of hamster cell was correlated with the prognosis of LGG patients, moreover, the researchers now have glial pain resistant CD8 + lymphocytes in the peripheral blood of glioma patients, these evidences suggest that gliomas have some immunogenicity, and it is closely related to the prognosis of LGG. Moreover, some studies have found that the immune system abnormalities can be found in LGG patients, including a decrease in the number of worker cells. T cell response capacity defect, early in the cell receptor transmembrane signal transduction impairment, other abnormal immune related cells including CD4 + and CD8 + T lymphocytes [16-18]. However, the specific mechanism of tumor immunity of tumor cells and immune cells in LGG has not been elucidated.
In this study, we performed a complex cluster analysis of LGG data samples from TCCA and CCGA databases. A special group (EC1) was found, whose prognosis was significantly worse than that of the other three groups (EC2-EC4) and combined with complex bioinformatics GSEA analysis methods showed that the genes of this groups were mainly involved in tumour immunity. The prognostic key genes SELL, ADCY8 and TLR4. Survival analysis showed that these genes were closely associated with patient prognosion and that these genes play an important role in tumor immune infiltration. The biological mechanism related to the prognosis of LGG gene will be further studied in the follow-up study.
Conclusion
In our study, the relationship between tumor and immune invasion was further explored by bioinformatics analysis, and SELL, ADCY8 and ADCY8 were multi-dimensionally analyzed by combining fine analysis, tumor tissue and medical big data. And the expression of TLR4 in LGG and its influence on the development of LGG, and the value of the above molecular markers in the diagnosis of LGG prognosis, in order to find new molecular markers of LGG and reveal the relationship between the expression of TLR4 and the development of LGG. LGG tumor immune mechanism provides theoretical and experimental basis.
Funding
This work was supported by the Scientific Research Project of Medical and Health Institutions of Longhua District, Shenzhen (No. 2020029), the Scientific Research Project of Medical and Health Institutions of Longhua District, Shenzhen (No. 2020108), the Scientific Research Project of Medical and Health Institutions of Longhua District, Shenzhen (No. 2021111).
Authors’ Contributions
Changpeng_wu is the major contributor in writing the manuscript, Hairen_zhou, Ling xiao, Ting tian provided suggestions to revise the manuscript, Shaoping_fan designed and revised the manuscript, All authors have reviewed the final manuscript.
Conflict of Interest
Authors declare no conflict of interest.
References
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, et al. (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114(2): 97-109.
- Zhang J, Stevens MF, Bradshaw TD (2012) Temozolomide: mechanisms of action, repair and resistance. Curr Mol Pharmacol 5(1): 102-114.
- Jones P S, Dunn G P, Nd B F, William T Curry, Fred H Hochberg, et al. (2013) Molecular genetics of low-grade gliomas: genomic alterations guiding diagnosis and therapeutic intervention. 11th annual Frye-Halloran Brain Tumor Symposium. Neurosurg Focus 34(2): E9.
- Pollack I F, Jakacki R, Butterfield L, Ronald L Hamilton, Ashok Panigrahy, et al. (2016) Immune responses and clinical outcome after glioma-associated antigen vaccination in children with recurrent low-grade gliomas. Neuro Oncol (4): 124.
- Lin W W, Ou G Y, Zhao W J (2021) Mutational profiling of low-grade gliomas identifies prognosis and immunotherapy-related biomarkers and tumour immune microenvironment characteristics. J Cell Mol Med 25(21): 10111-10125.
- Liang, RuofeiChen, NiLi, MaoWang, XiangMao, et al. (2018) Significance of systemic immune-inflammation index in the differential diagnosis of high- and low-grade gliomas. Clin neurol neurosurg 164: 50-52.
- Teng W, Wang L, Xue W, Chao Guan (2009) Activation of TLR4-mediated NFkappaB signaling in hemorrhagic brain in rats. Mediators Inflamm 2009(1): 473276.
- Mallikarjuna P, Wai M C, Beng K Y (2013) Should a Toll-like receptor 4 (TLR-4) agonist or antagonist be designed to treat cancer? TLR-4: its expression and effects in the ten most common cancers. OncoTargets Ther 6: 1573-1587.
- Prasad K N, Cole W C, Yan X D, B Kumar, A Hanson, J E Prasad (2003) Defects in cAMP-pathway may initiate carcinogenesis in dividing nerve cells: A review. Apoptosis 8(6): 579-586.
- Wilkerson Matthew, D Hayes (2010) ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26(12): 1572-1573.
- Aravind Subramanian, Heidi Kuehn, Joshua Gould, Pablo Tamayo, Jill P Mesirov (2007) GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 23(23): 3251-3253.
- Lin W W, Ou G Y, Zhao W J (2021) Mutational profiling of low-grade gliomas identifies prognosis and immunotherapy-related biomarkers and tumour immune microenvironment characteristics. J Cell Mol Med 25(21): 10111-10125.
- Zijian Zhou, Jinhong Wei, Bin Lu, Wenbo Jiang , Yue Bao, et al. (2022) Comprehensive Characterization of Pyroptosis Patterns with Implications in Prognosis and Immunotherapy in Low-Grade Gliomas. Front Genet 12: 763807.
- Chen SS (2012) The treatment of adult low-grade glioma. Chinese J Neuro Oncol 60(3): 90-108.
- Luo H, Huang K, Tao C, Wu M, Ye M, et al. (2020) The Prognostic Value of Risk Score Based on Immunogenenomic Landscape Analysis in Glioma.
- Learn CA, Schmittling RJ, Fecci PE, Mitchell DA, Archer GE, et al. (2005) Expression profiling of CD4+ and CD8+ T cells in malignant glioma. [C]//Aacr/nci/eortc International Conference on Molecular Targets & Cancer.
- Ge Jing, Zhao Lina, Li Guanggang, White Jason, Song, et al. (2017) Cytotoxic CD4(+) T cells are correlated with better prognosis in Han Chinese grade II and grade III glioma subjects and are suppressed by PD-1 signaling. Int J Neurosci 127(5): 386-395.
- Genestie C, Descamps Dudez, et al. (2016) Heterogeneity in PD-L1 expression and CD8+infiltrates in low grade versus high grade serous ovarian carcinomas. Virchows Archiv: an international journal of pathology pp. 469.




 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.